Cleaved PARP1 Rabbit mAb [YJZT]Cat NO.: A37266
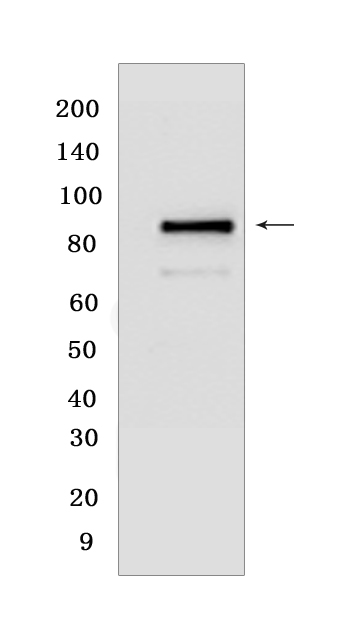
Western blot(SDS PAGE) analysis of extracts from Jurkat cells .Using Cleaved PARP1Rabbit mAb [YJZT] at dilution of 1:1000 incubated at 4℃ over night.
Product information
Protein names :PARP1,ADPRT,PPOL,PARP1_HUMAN,Poly [ADP-ribose] polymerase 1 ADP-ribosyltransferase 1)
UniProtID :P09874
MASS(da) :113,084
MW(kDa) :85 kDa
Form :Liquid
Purification :Protein A purification
Host :Rabbit
Isotype :IgG
sensitivity :Endogenous
Reactivity :Human
- ApplicationDilution
- 免疫印迹(WB)1:1000-2000
- The optimal dilutions should be determined by the end user
Specificity :Antibody is produced by immunizing animals with a synthetic peptide at the sequence of human Cleaved PARP1
Storage :Antibody store in 10 mM PBS, 0.5mg/ml BSA, 50% glycerol. Shipped at 4°C. Store at-20°C or -80°C. Products are valid for one natural year of receipt.Avoid repeated freeze / thaw cycles.
WB Positive detected :Jurkat cells
Function : Poly-ADP-ribosyltransferase that mediates poly-ADP-ribosylation of proteins and plays a key role in DNA repair (PubMed:17177976, PubMed:18172500, PubMed:19344625, PubMed:19661379, PubMed:21680843, PubMed:23230272, PubMed:25043379, PubMed:33186521, PubMed:32028527, PubMed:26344098). Mediates glutamate, aspartate, serine or tyrosine ADP-ribosylation of proteins: the ADP-D-ribosyl group of NAD(+) is transferred to the acceptor carboxyl group of target residues and further ADP-ribosyl groups are transferred to the 2'-position of the terminal adenosine moiety, building up a polymer with an average chain length of 20-30 units (PubMed:7852410, PubMed:9315851, PubMed:19764761, PubMed:25043379, PubMed:28190768, PubMed:29954836). Serine ADP-ribosylation of proteins constitutes the primary form of ADP-ribosylation of proteins in response to DNA damage (PubMed:33186521). Mainly mediates glutamate and aspartate ADP-ribosylation of target proteins in absence of HPF1 (PubMed:19764761, PubMed:25043379). Following interaction with HPF1, catalyzes serine ADP-ribosylation of target proteins,HPF1 conferring serine specificity by completing the PARP1 active site (PubMed:28190768, PubMed:29954836, PubMed:33186521, PubMed:32028527). Also catalyzes tyrosine ADP-ribosylation of target proteins following interaction with HPF1 (PubMed:30257210, PubMed:29954836). PARP1 initiates the repair of DNA breaks: recognizes and binds DNA breaks within chromatin and recruits HPF1, licensing serine ADP-ribosylation of target proteins, such as histones, thereby promoting decompaction of chromatin and the recruitment of repair factors leading to the reparation of DNA strand breaks (PubMed:17177976, PubMed:18172500, PubMed:19344625, PubMed:19661379, PubMed:23230272, PubMed:27067600). In addition to base excision repair (BER) pathway, also involved in double-strand breaks (DSBs) repair: together with TIMELESS, accumulates at DNA damage sites and promotes homologous recombination repair by mediating poly-ADP-ribosylation (PubMed:26344098, PubMed:30356214). Mediates the poly(ADP-ribosyl)ation of a number of proteins, including itself, APLF and CHFR (PubMed:17396150, PubMed:19764761). In addition to proteins, also able to ADP-ribosylate DNA: catalyzes ADP-ribosylation of DNA strand break termini containing terminal phosphates and a 2'-OH group in single- and double-stranded DNA, respectively (PubMed:27471034). Required for PARP9 and DTX3L recruitment to DNA damage sites (PubMed:23230272). PARP1-dependent PARP9-DTX3L-mediated ubiquitination promotes the rapid and specific recruitment of 53BP1/TP53BP1, UIMC1/RAP80, and BRCA1 to DNA damage sites (PubMed:23230272). Acts as a regulator of transcription: positively regulates the transcription of MTUS1 and negatively regulates the transcription of MTUS2/TIP150 (PubMed:19344625). Plays a role in the positive regulation of IFNG transcription in T-helper 1 cells as part of an IFNG promoter-binding complex with TXK and EEF1A1 (PubMed:17177976). Involved in the synthesis of ATP in the nucleus, together with NMNAT1, PARG and NUDT5 (PubMed:27257257). Nuclear ATP generation is required for extensive chromatin remodeling events that are energy-consuming (PubMed:27257257)..
Subcellular locationi :Nucleus. Nucleus, nucleolus. Chromosome.
IMPORTANT: For western blots, incubate membrane with diluted primary antibody in 1% w/v BSA, 1X TBST at 4°C overnight.