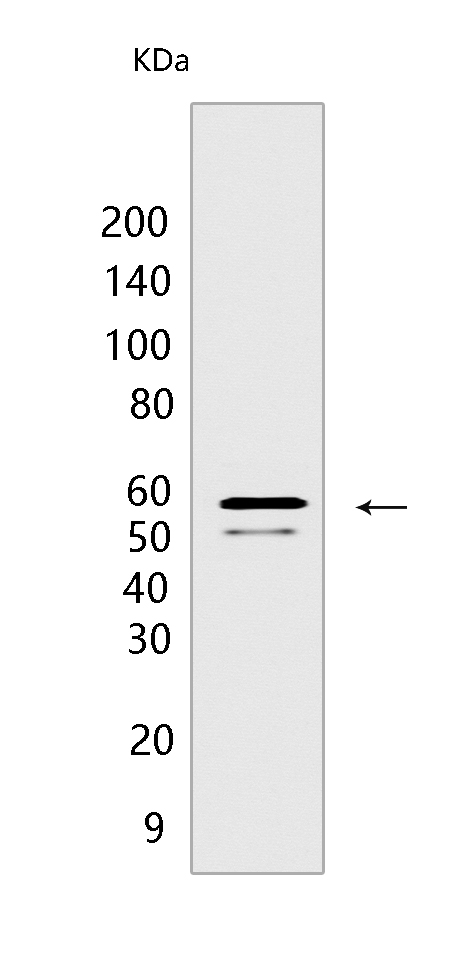
RAD23B Mouse mAb[T3VU]Cat NO.: A83850
Western blot(SDS PAGE) analysis of extracts from LNCaP cells.Using RAD23B Mouse mAb IgG [T3VU] at dilution of 1:1000 incubated at 4℃ over night.
Product information
Protein names :RAD23B,RD23B_HUMAN,UV excision repair protein RAD23 homolog B
UniProtID :P54727
MASS(da) :43,171
MW(kDa) :58kDa
Form :Liquid
Purification :Protein A purification
Host :Mouse
Isotype :IgG
sensitivity :Endogenous
Reactivity :Human,Mouse,Rat
- ApplicationDilution
- 免疫印迹(WB)1:1000-2000
- 免疫组化(IHC)1:100
- 免疫荧光(ICC/IF) 1:100,
- The optimal dilutions should be determined by the end user
Specificity :Antibody is produced by immunizing animals with a synthetic peptide of human RAD23B.
Storage :Antibody store in 10 mM PBS, 0.5mg/ml BSA, 50% glycerol. Shipped at 4°C. Store at-20°C or -80°C. Products are valid for one natural year of receipt.Avoid repeated freeze / thaw cycles.
WB Positive detected :LNCaP cells
Function : Multiubiquitin chain receptor involved in modulation of proteasomal degradation. Binds to polyubiquitin chains. Proposed to be capable to bind simultaneously to the 26S proteasome and to polyubiquitinated substrates and to deliver ubiquitinated proteins to the proteasome. May play a role in endoplasmic reticulum-associated degradation (ERAD) of misfolded glycoproteins by association with PNGase and delivering deglycosylated proteins to the proteasome., Involved in global genome nucleotide excision repair (GG-NER) by acting as component of the XPC complex. Cooperatively with CETN2 appears to stabilize XPC. May protect XPC from proteasomal degradation., The XPC complex is proposed to represent the first factor bound at the sites of DNA damage and together with other core recognition factors, XPA, RPA and the TFIIH complex, is part of the pre-incision (or initial recognition) complex. The XPC complex recognizes a wide spectrum of damaged DNA characterized by distortions of the DNA helix such as single-stranded loops, mismatched bubbles or single-stranded overhangs. The orientation of XPC complex binding appears to be crucial for inducing a productive NER. XPC complex is proposed to recognize and to interact with unpaired bases on the undamaged DNA strand which is followed by recruitment of the TFIIH complex and subsequent scanning for lesions in the opposite strand in a 5'-to-3' direction by the NER machinery. Cyclobutane pyrimidine dimers (CPDs) which are formed upon UV-induced DNA damage esacpe detection by the XPC complex due to a low degree of structural perurbation. Instead they are detected by the UV-DDB complex which in turn recruits and cooperates with the XPC complex in the respective DNA repair. In vitro, the XPC:RAD23B dimer is sufficient to initiate NER,it preferentially binds to cisplatin and UV-damaged double-stranded DNA and also binds to a variety of chemically and structurally diverse DNA adducts. XPC:RAD23B contacts DNA both 5' and 3' of a cisplatin lesion with a preference for the 5' side. XPC:RAD23B induces a bend in DNA upon binding. XPC:RAD23B stimulates the activity of DNA glycosylases TDG and SMUG1.
Subcellular locationi :Nucleus. Cytoplasm.
IMPORTANT: For western blots, incubate membrane with diluted primary antibody in 1% w/v BSA, 1X TBST at 4°C overnight.