SNF2L Rabbit mAb [EUOL]Cat NO.: A44469
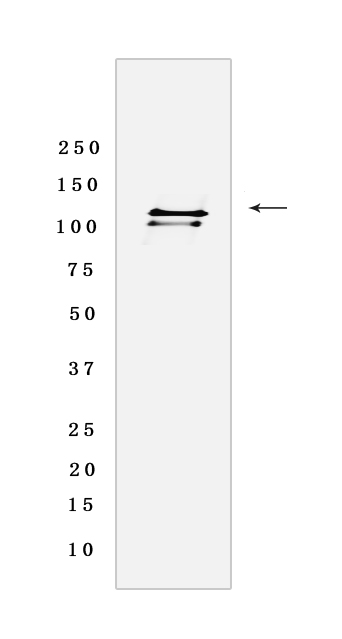
Western blot analysis of extracts from Hela cells lyastes.using SNF2L Rabbit mAb [EUOL] at dilution of 1:1000 incubated at 4℃ over night
Product information
Protein names :SMARCA1,SNF2L,SNF2L1,SMCA1_HUMAN,Probable global transcription activator SNF2L1
UniProtID :P28370
MASS(da) :122,605
MW(kDa) :123 kDa
Form :Liquid
Purification :Protein A purification
Host :Rabbit
Isotype :IgG
sensitivity :Endogenous
Reactivity :Human
- ApplicationDilution
- 免疫印迹(WB)1:1000-2000
- 免疫组化(IHC)1:100,
- 免疫荧光(ICC/IF)1:100,
- The optimal dilutions should be determined by the end user
Specificity :Antibody is produced by immunizing animals with a synthetic peptide of Human SNF2L.
Storage :Antibody store in 10 mM PBS, 0.5mg/ml BSA, 50% glycerol. Shipped at 4°C. Store at-20°C or -80°C. Products are valid for one natural year of receipt.Avoid repeated freeze / thaw cycles.
WB Positive detected :Hela cells lyastes
Function : [Isoform 1]: Catalytically inactive when either DNA or nucleosomes are the substrate and does not possess chromatin-remodeling activity (PubMed:15310751, PubMed:28801535). Acts as a negative regulator of chromatin remodelers by generating inactive complexes (PubMed:15310751).., [Isoform 2]: Helicase that possesses intrinsic ATP-dependent chromatin-remodeling activity (PubMed:15310751, PubMed:14609955, PubMed:15640247, PubMed:28801535). ATPase activity is substrate-dependent, and is increased when nucleosomes are the substrate, but is also catalytically active when DNA alone is the substrate (PubMed:15310751, PubMed:14609955, PubMed:15640247). Catalytic subunit of ISWI chromatin-remodeling complexes, which form ordered nucleosome arrays on chromatin and facilitate access to DNA during DNA-templated processes such as DNA replication, transcription, and repair (PubMed:15310751, PubMed:14609955, PubMed:15640247, PubMed:28801535). Within the ISWI chromatin-remodeling complexes, slides edge- and center-positioned histone octamers away from their original location on the DNA template (PubMed:28801535). Catalytic activity and histone octamer sliding propensity is regulated and determined by components of the ISWI chromatin-remodeling complexes (PubMed:28801535). The BAZ1A-, BAZ1B-, BAZ2A- and BAZ2B-containing ISWI chromatin-remodeling complexes regulate the spacing of nucleosomes along the chromatin and have the ability to slide mononucleosomes to the center of a DNA template (PubMed:28801535). The CECR2- and RSF1-containing ISWI chromatin-remodeling complexes do not have the ability to slide mononucleosomes to the center of a DNA template (PubMed:28801535). Within the NURF-1 and CERF-1 ISWI chromatin remodeling complexes, nucleosomes are the preferred substrate for its ATPase activity (PubMed:14609955, PubMed:15640247). Within the NURF-1 ISWI chromatin-remodeling complex, binds to the promoters of En1 and En2 to positively regulate their expression and promote brain development (PubMed:14609955). May promote neurite outgrowth (PubMed:14609955). May be involved in the development of luteal cells (PubMed:16740656)..
Tissue specificity :[Isoform 1]: Mainly expressed in non-neuronal tissues such as lung, breast, kidney, and ovary..,[Isoform 2]: Expressed in lung, breast, kidney, ovary, skeletal muscle and brain..
Subcellular locationi :Nucleus.
IMPORTANT: For western blots, incubate membrane with diluted primary antibody in 1% w/v BSA, 1X TBST at 4°C overnight.